Crop genetic resources refer to the diversity of traditional landraces and modern cultivated varieties, including their crop wild relatives. The number of cultivated crops has been drastically reduced by the intensification of agriculture since the 20th century. Today, around 30 species are used to satisfy 90% of humanity's needs, whereas 100 species were used at the beginning of the 20th century (Gepts, 2006). In this context of crop genetic erosion, in situ and ex situ conservation of a wide range of crop diversity is essential to ensure food security and to face global changes (FAO, 2010; Khoury et al, 2014; FAO, 2020).
The ex situ approach involves safeguarding crop diversity outside of its native environment, typically within conservatories, or specialized infrastructures such as seedbanks. The primary goal is to conserve and propagate crop genetic resources and make them available for research, breeding and cultivation. This is particularly crucial for neglected and underutilized species (NUS) that received limited scientific attention (Stamp et al, 2012; Hunter et al, 2019; Ulian et al, 2020), whereas they could be used to face global changes and improve the quality and sustainability of food production (Ulian et al, 2020).
Describing and characterizing NUS accessions that are preserved in ex situ collections is essential for their management and sustainable use. Accurate documentation of accessions enables informed decisions to be made on conservation, research, breeding and potential use (Weise et al, 2020). However, this conservation approach requires the availability of accurate passport data, notably to avoid any taxonomic misidentification (Guzzon et al, 2018) or geographic location errors, which can introduce spatial bias into databases and distort large-scale biodiversity analyses (Beck et al, 2014).
Among NUS, fonio is a key cereal, native to West Africa, with valuable nutritional and agronomic qualities. Fonio is highly adapted to harsh environmental conditions and plays a crucial role in food security within developing economies. The potential of fonio has earned it recognition by the Value Addition in Cereal Systems (VACS) initiative as a top cereal for West Africa (Karl et al, 2024). The accuracy of passport data is particularly critical in the case of fonio. Fonio comprises two similar species with tiny seeds, both grown in West Africa, sometimes in the same localities; identifying the two species is not obvious. The most common, Digitaria exilis Stapf, is known as white fonio and is cultivated in an area stretching from Senegal to Nigeria. The second, Digitaria iburua Stapf, is named black fonio and its distribution is limited to northern Nigeria, Togo and Benin (Animasaun et al, 2018). The black and white denomination for fonio refers to the colour of the seed husks, which are likely to be more intensely dark brown for D. iburua than D. exilis (Adoukonou-Sagbadja A-H, 2010) (Figure 1). However, this criterion can vary within the two species, leading to confusion. The variability of this trait, plus the fact that fonio species are sometimes not distinguished by their common name (Blench, 2016), can lead to misidentification in collections.

Improving the accuracy of genebank passport data concerning the identification of D. exilis and D. iburua is a key issue that needs to be overcome to preserve fonio genetic resources, and make their adaptive potential available to farmers. Fonio identification could be based on vegetative, floral or spikelet characteristics, or molecular markers. For example, the growth habits of white and black fonio differ (Figure 2), but using this trait as an identification criterion requires seed growing, as well as access to large-scale cultivation areas or costly infrastructures. In addition, genebank collections often contain only limited seed samples for some fonio accessions. On the other hand, genetic identification methods, based on microsatellite genotyping or genome sequencing, require small samples (Mondini et al, 2009). All these methods are expensive, destructive and time-consuming.
The aim of this work was to develop a low-cost, non-destructive and rapid method, based on seed morphology, in order to assign fonio accessions to either white or black fonio species. To date, the seed morphometrics approach has never been applied to fonio crops. Such an approach is highly relevant for fonio genebank collections to improve the quality of associated passport data and enhance the value of these collections.

Plant material and sampling
Fonio accessions are maintained in the seed collection in Montpellier, France, at the GAMéT Resource Centre (ARCAD) and the French National Research Institute for Sustainable Development (IRD) and in national genebank collections across West Africa. These collections of seeds (paddy grains), sometimes in small amounts (less than one gram), have been built up since 1977 thanks to collection missions to farmers in the areas of origin and thanks to partnerships between French and African research institutions involved in various research projects.
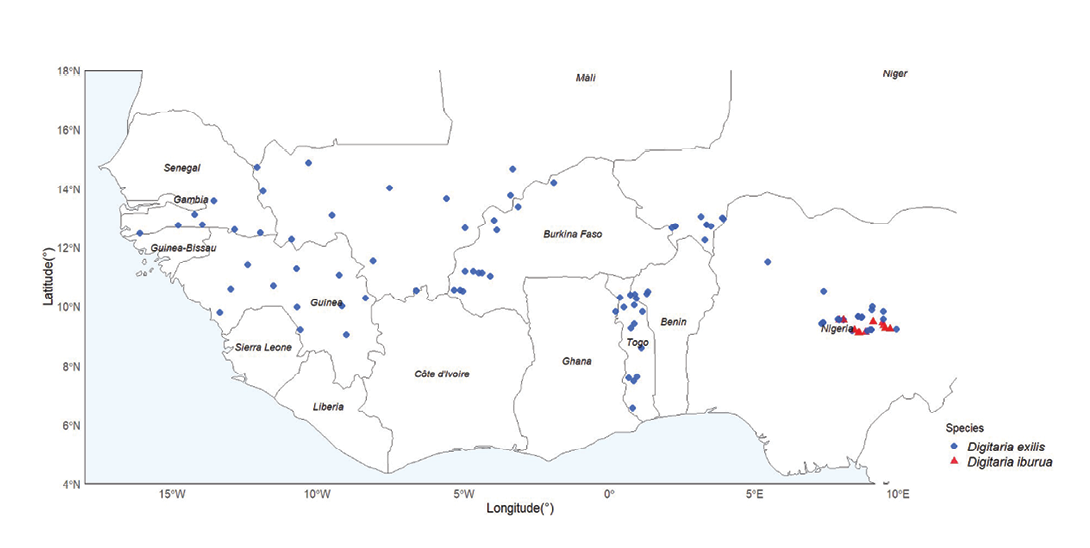
A sample of 118 accessions of D. exilis and D. iburua (98 and 20 respectively, Supplemental Table 1) previously sequenced in Abrouk et al (2020) and Kaczmarek et al (2025) was selected to maximize geographical coverage (Figure 3) and thus climatic diversity. Only accessions whose species had been genetically validated were selected.
For each accession, one seed sample was prepared in a 2ml microtube, weighing between 110mg and 151.4mg. The variability in sample mass was linked to the quantity of seeds available and to the difficulty of handling very small seeds (in the millimetre range, Figure 1).
Seed image analysis
For each accession, the seed sample was poured and carefully laid on a 13.5 × 10.5cm surface of a flatbed scanner (Epson Expression 10000XL) to be scanned on a green background (Canson-C200040066). An average of 232 seeds per accession was scanned. In total, 27,345 seeds were analyzed. The images were saved in .tif format (800dpi resolution).
The images were analyzed with the Rigatoni v.0.9.3 R package (Rami, 2022), based on the EBImage R package. The Rigatoni package was designed to analyze seed images acquired by scanning. In contrast to the colour of background pixels, the algorithm detects objects in an image and characterizes their size, shape and colour, with a total of 27 descriptors. Each seed was individually cropped in the image using the krnel function.
Preliminary tests were carried out to calibrate the seed detection algorithm and define size and colour thresholds to avoid the detection of artefacts. It was observed that fonio seeds cannot be smaller than 500px (minimum size threshold for an object), nor larger than 2,000px (maximum size threshold). To find the seeds in the image, the hue range of the green background was set between 68° and 92° and the brightness threshold was set at 0.001.
Dimensions in pixels were converted to tenths-of-a-millimeter (tmm), and the area was converted from pixels to square tenths-of-a-millimetre (tmm2). The hexadecimal colour code was also determined from each seed’s cropped image. For data analysis, this colour code in RGB components was then converted into H, S and V components with the colorspace v2.0-3 R package (Zeileis et al, 2020). The advantage of the HSV colour system is that it is based on components perceived by humans to describe colours: hue (tint or predominant colour), saturation (colour intensity) and value (brightness), allowing intuitive interpretation of colour variations (Hema et al, 2019).
Preliminary exploratory data analysis
A graphical exploratory analysis was performed on the values measured per seed, and revealed distributions that were sometimes highly asymmetrical with very extreme and therefore suspicious values. An automatic extreme values filter was applied with the boxplot.stats() function, using the interquartile range (IQR) and a whiskers coefficient of 3 times this length to eliminate only highly improbable values. For each descriptor and accession, any value below Q1 - 3IQR, or above Q3 + 3IQR was considered an extreme outlier, with Q1 as the lower quartile and Q3 as the upper quartile. Seeds with at least one descriptor presenting an extreme outlier value were excluded from the data. For further data analysis, the morphometric values measured per seed were summarized, for each accession, by their median value.
Morphometric and colour descriptors
An exploratory analysis of the 27 descriptors was carried out to assess their variation and reduce their number in the event of strong correlations. A graphical method was used to explore the relationship between descriptors in pairs (matrix of graphs, not shown), initially considering separately each category: size, shape, pixel intensity, colour, contour (see Table 1 for descriptor details). Thousand-grain weight was added as a usual descriptor for cereals. Descriptors were selected in such a way as to retain only those descriptors that made sense, i.e. those that provided specific and easily understandable information on the sample variability. In the case of highly correlated variables, the selected variable was the one that made more sense. For example, s.area (seed area) and s.radius.mean (mean seed radius, Table 1) were highly correlated (Pearson correlation value 0.99), hence seed area, which made more sense than s.radius.mean, was selected. Variables representing statistical dispersion parameters such as standard deviation, median absolute deviation, or quantile, were not selected. All variables that were complementary to each other, such as the H, S and V colour parameters, were selected.
Statistical analysis of morphometric data
Descriptive statistics were carried out using the stat.desc() function of the pastecs v.1.3.21 R package (Grosjean et al, 2018) in order to characterize the two species. Histograms of the median values per accession for each species (Supplemental Figure 1) showed non-normal (skewed and/or over-spread) distributions for most morphometric variables. These data were not suitable for the application of a t-test, and the two fonio species were compared by the Mann-Whitney-Wilcoxon rank test (wilcox.test() function). Morphometric diversity of the 118 accessions was explored using principal component analysis (PCA). PCA was performed on the seven seed descriptors (Table 1), using the PCA() function in the FactoMineR v.2.6 R package (Lê et al, 2008).
Table 1. Characteristics of the morphometric and colour descriptors. *, descriptors selected for the study; tmm, tenths-of-a-millimeter; tmm2, square tenths-of-a-millimeter; sqrt, square root.
|
Descriptor |
Characteristics |
|
Seed size (provided by Rigatoni package) |
|
|
bbox.width* |
Object bounding box width (in pixels, converted to tmm) measuring seed width |
|
bbox.height* |
Object bounding box height (in pixels, converted to tmm) measuring seed length |
|
s.area* |
Area, number of pixels in the shape (converted to tmm2) |
|
s.perimeter |
Perimeter, number of pixels in the boundary of the object (converted to tmm) |
|
s.radius.mean |
Mean radius (in pixels), average radius value from the centre of shape to boundary (converted to tmm) |
|
s.radius.sd |
Standard deviation of the radius values (in pixels) |
|
s.radius.max |
Max radius (in pixels), largest radius value from the centre of shape to boundary (converted to tmm) |
|
s.radius.min |
Min radius (in pixels), shortest radius value from the centre of shape to boundary (converted to tmm) |
|
Seed shape (provided by Rigatoni package) |
|
|
m.eccentricity* |
Elliptical eccentricity, values ranging from 0 (perfect circle) to 1 (straight-line). Calculated with the longest axis (majoraxis) and the shortest axis (minoraxis) of the best-fitting ellipse: sqrt(1-minoraxis2/majoraxis2). |
|
m.majoraxis |
Largest axis of the best-fitting ellipse (in pixels, converted to tmm) |
|
m.cx, m.cy |
Centre of the best-fitting ellipse coordinates (in pixels) |
|
m.theta |
Object angle (in radians) |
|
Pixels intensity (provided by Rigatoni package) |
|
|
b.mean |
Average of pixel intensity in the shape |
|
b.sd |
Standard deviation of pixel intensity in the shape |
|
b.mad |
Median absolute deviation of pixel intensity in the shape |
|
b.q (b.q001, b.q005, b.q05, b.q095, b.q099) |
Quantile intensity of pixel intensity in the shape |
|
Seed contour (provided by Rigatoni package) |
|
|
poi.x, poi.y |
Pole of inaccessibility coordinates, coordinates of the point farthest away from the boundary of the object |
|
poi.dist |
Longest distance to the boundary of the object (in pixels, converted to tmm) |
|
Seed colour (obtained by converting the RGB code provided by Rigatoni package) |
|
|
H* |
Hue (in degrees), predominant colour or tint, (values ranging from 0 to 360°) |
|
S* |
Saturation, intensity of colour pigmentation (values ranging from 0 to 1) |
|
V* |
Value, brightness of the colour (values ranging from 0 to 1) |
|
Thousand-grain weight (calculated by accession) |
|
|
TGW* |
Thousand seed weight (in grams) |
Results
The objective of this study was to develop an affordable, non-destructive and rapid method based on seed morphology to categorize fonio accessions into white or black fonio species.
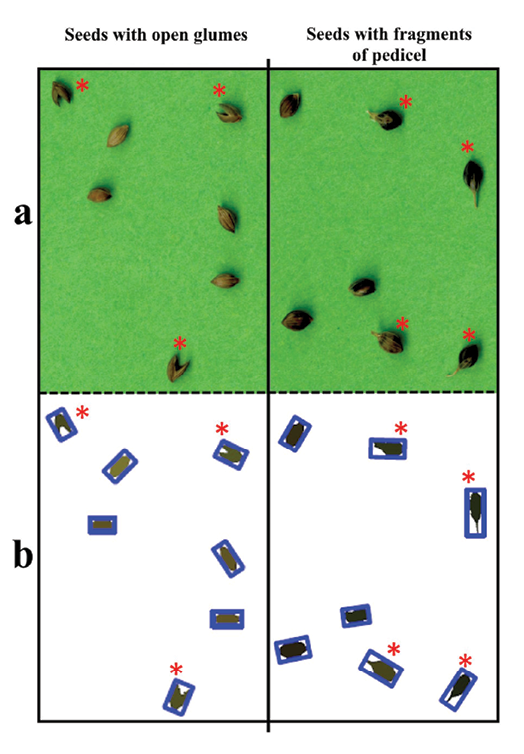
The 3IQR method discarded 1.1% of the 27,345 seeds analyzed. Nine accessions had no outliers. For the remaining 109 accessions, the percentage of outliers varied from 0.4% to 8.8% (Supplemental Table 2). No link could be established between the percentage of outliers and the parameters structuring the sampling design (species, country of origin and collection date).
Figure 4 shows an example of how outliers were detected for seeds with attached pedicels and seeds with open glumes.
Morphometrics description of black and white fonio
D. iburua and D. exilis differed significantly for all variables (Wilcoxon test at the p-value threshold < 0.05). It can be noted that seed area (s.area) and seed width (bbox.width) showed no overlap at all between the two species for our sample of accessions (Supplemental Figure 1). The results presented below are based on the descriptors' median values.
D. iburua seeds were significantly wider (+19%), longer (+13%) and heavier (31%) than D. exilis seeds (Wilcoxon test, p < 0.001, Supplemental Figure 1). D. iburua seeds were 19.1tmm long (bbox.height) and 11.1tmm wide (bbox.width), while D. exilis seeds were 16.9tmm long and 9.4tmm wide. The thousand-grain weight was equal to 0.71g for D. iburua and 0.54g for D. exilis (Table 2).
For seed width (bbox.width), the range of variation between the minimum and the maximum values showed a clear demarcation between the two species (Supplemental Figure 1). Indeed, seed width varied for D. exilis from 8.2 tmm to 10.1tmm as compared to 10.5tmm to 12.0tmm for D. iburua accessions (Table 2). Seed area (s.area) values also highly differed (Wilcoxon test, p < 0.001), revealing a distinct separation between the two species. For D. exilis, seed area varied from 90.6tmm2 to 133.7tmm2 whereas for D. iburua, seed area varied from 139.8tmm2 to 162.7tmm2 (Table 2). These results confirmed that D. iburua seeds were significantly bigger than those of D. exilis.
Furthermore, m.eccentricity values were significantly (Wilcoxon test, p < 0.05) higher for D. exilis (0.82) than for D. iburua (0.80). The distribution of values for each species (Supplemental Figure 1) showed a second peak at high values of m.eccentricity (around m.eccentricity = 0.83), indicating that some D. exilis accessions had, on average, more elongated seeds.
Colour analysis
Hue values were significantly different (Wilcoxon test, p < 0.001) between D. iburua (H = 51.9°) and D. exilis (H= 56.4°). Both values were in the yellow range, but D. iburua seeds had a warmer yellow-red hue than D. exilis seeds whose hue was closer to pure yellow (Supplemental Figure 2). Moreover, colour brightness (V) values were significantly (Wilcoxon test, p < 0.001) lower for D. iburua (V = 0.21) than for D. exilis (V = 0.32); reversely, colour saturation (S) values were significantly (Wilcoxon test, p < 0.001) higher for D. iburua (S = 0.89) than for D. exilis (S= 0.77) (Table 2). Those contrasted V and S values, associated with H values correspond to the contrasted colour of the seed husks, which are dark brown for D. iburua and light brown for D. exilis (Figure 1).
Morphometric diversity
The PCA analysis revealed a clear distinction between white and black fonio (Figure 5). The first four principal components covered 99% of the total variance (Supplemental Figure 3).
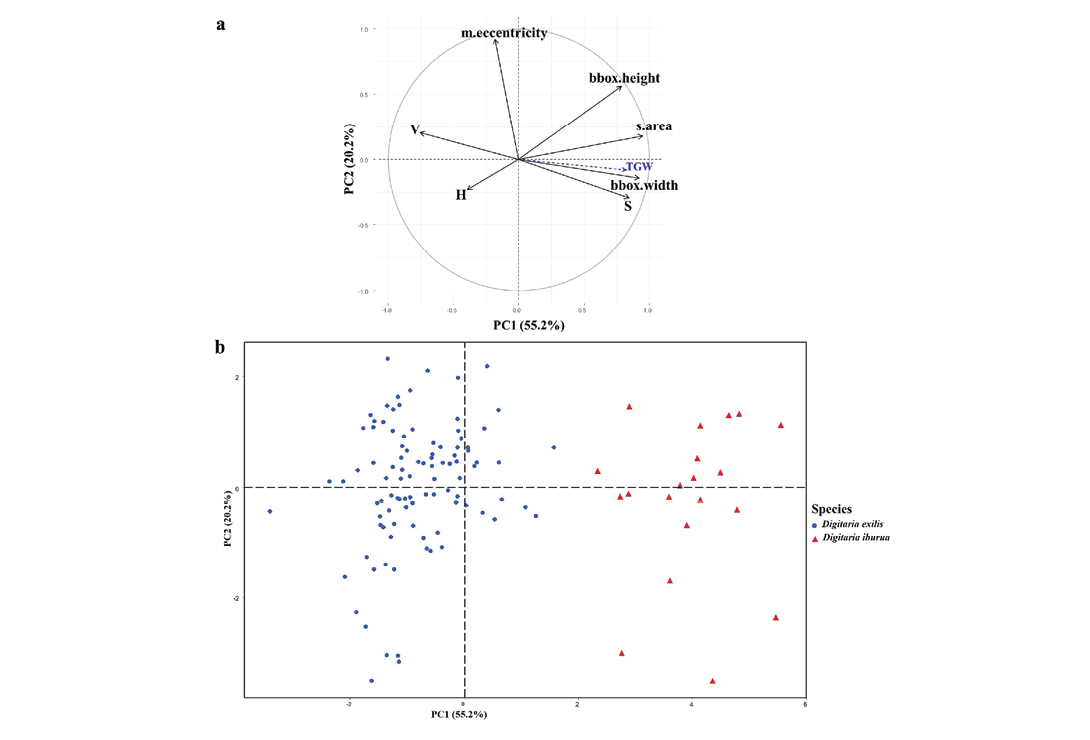
Figure 5. Principal component analysis carried out on the seed morphometric measurements of the 118 fonio accessions (n = 98 for Digitaria exilis and n = 20 for Digitaria iburua). a, Correlation circle for the first two principal components, where bbox.width correspond to width, bbox.height to length, S to saturation of colour, H to hue of colour, V to brightness of colour, m.eccentricity to eccentricity, s.area to seed area, and TGW to thousand-grain weight. The supplementary variable TGW was coloured in blue. b, Scatterplot of the 118 fonio accessions projected on the first two principal components plane.
Table 2. Descriptive statistics for size and shape analysis of each fonio species. N, number of accessions.
|
Variables |
Minimum value (Min) |
Maximum value (Max) |
Median |
Mean |
Standard deviation (SD) |
Coefficient of variation (CV, %) |
|
|
Digitaria exilis (N = 98) |
s.area (tmm2) |
90.63 |
133.72 |
113.28 |
112.80 |
71.91 |
7.5 |
|
bbox.width (tmm) |
8.24 |
10.09 |
9.37 |
9.34 |
0.10 |
3.4 |
|
|
bbox.height (tmm) |
14.34 |
19.05 |
16.86 |
16.85 |
0.95 |
5.8 |
|
|
m.eccentricity |
0.74 |
0.86 |
0.82 |
0.82 |
0.00 |
2.9 |
|
|
H (°) |
53.81 |
62.56 |
56.40 |
56.41 |
1.89 |
2.4 |
|
|
S |
0.65 |
0.87 |
0.77 |
0.76 |
0.00 |
5.5 |
|
|
V |
0.22 |
0.48 |
0.32 |
0.32 |
0.00 |
11.6 |
|
|
TGW (g) |
0.31 |
0.70 |
0.54 |
0.53 |
0.00 |
12.2 |
|
|
Digitaria iburua (N = 20) |
s.area (tmm2) |
139.82 |
162.70 |
153.23 |
152.86 |
46.02 |
4.4 |
|
bbox.width (tmm) |
10.54 |
11.96 |
11.11 |
11.14 |
0.14 |
3.3 |
|
|
bbox.height (tmm) |
17.89 |
20.32 |
19.11 |
19.17 |
0.61 |
4.1 |
|
|
m.eccentricity |
0.74 |
0.84 |
0.80 |
0.80 |
0.00 |
3.4 |
|
|
H (°) |
48.24 |
71.71 |
51.86 |
53.63 |
35.54 |
11.1 |
|
|
S |
0.80 |
0.94 |
0.89 |
0.89 |
0.00 |
3.7 |
|
|
V |
0.11 |
0.37 |
0.21 |
0.23 |
0.01 |
35.3 |
|
|
TGW (g) |
0.59 |
0.88 |
0.71 |
0.73 |
0.01 |
9.8 |
The first principal component (PC1, 55.2% of the total variance) opposed seed size parameters and saturation (S) of the seed colour (positive values), with brightness (V) of the seed colour (negative values, Figure 5a). Seed size refers to seed area (s.area), seed width (bbox.width) and seed length (bbox.height), which were among the most influential characters on PC1 (Supplemental Table 3). Variability of s.area and bbox.width variables was almost entirely represented by PC1 (cos2: resp. 90% and 85%, Supplemental Table 3). The PC2 (20.2% of the total variance) was mainly related to the m.eccentricity variable (contribution: 60.7%), which is a component of the seed shape, and to a lesser extent to seed length (bbox.height, contribution: 22.5%). The variability of the m.eccentricity variable is almost entirely represented by PC2 (cos2: 86%). PC3 (13.9% of the total variance) was mainly related to the tint (H, contribution: 80%). For PC4, which accounted for less than 10% of total variability, the brightness (V) of the seed colour was the most influential character (contribution: 37%) (Supplemental Table 3).
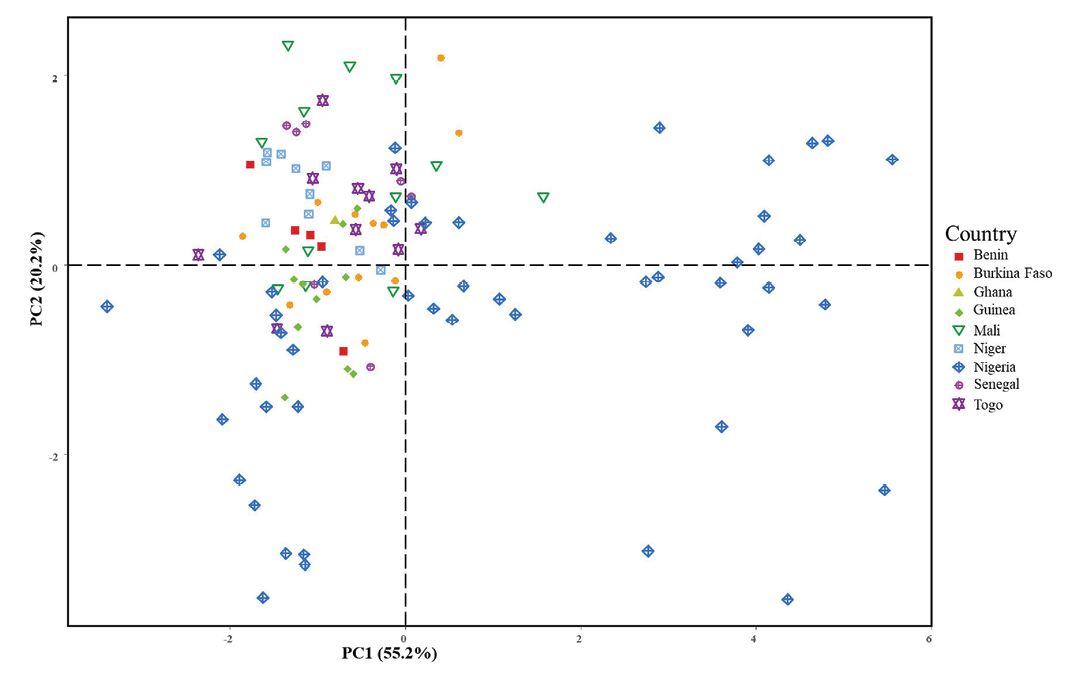
The first axis (Figure 5b) completely differentiated the two fonio species, with black fonio (right) seeds characterized by higher values of area, width, colour saturation (S) and height, and lower values of brightness (V), compared to white fonio (left) with the opposite characteristics. Independent of this clear structure separating the species on the first axis, the second axis showed, within each species, a gradient of variation in seed shape (m.eccentricity), from the least to the most elongated. Illustration of accessions by their country of origin (Figure 6) revealed that the roundest seeds (bottom of axis 2) originated from Nigeria only. Moreover, the Nigerian accessions were projected over the same range along this axis for both species. On the opposite side and on the top of axis 2, the white fonio accessions with the most tapered seeds mainly originated from Mali and Senegal.
The first factorial plane concentrated 75% of the total variability and made it possible to characterize the morphometric differences between the seeds of the two species. The next PCs, calculated on the remaining variability, did not appear to be informative. They showed either particularities for some accessions (PC3), or more continuous variations which could not be linked to explanatory factors and therefore could not be interpreted (PC4).
Differentiation of the two species with only one morphometric descriptor
Figure 7 clearly showed that the seed width alone perfectly separated the two fonio species. Descriptive statistics (Table 2) specified the limiting values observed for these data: up to a value of seed width equal to 10.09tmm for D. exilis (n = 98) and from 10.54tmm for D. iburua (n = 20). In addition, the seed area parameter, which is strongly dependent on seed width and height, was also a differentiating trait between the two species (Supplemental Figure 4).
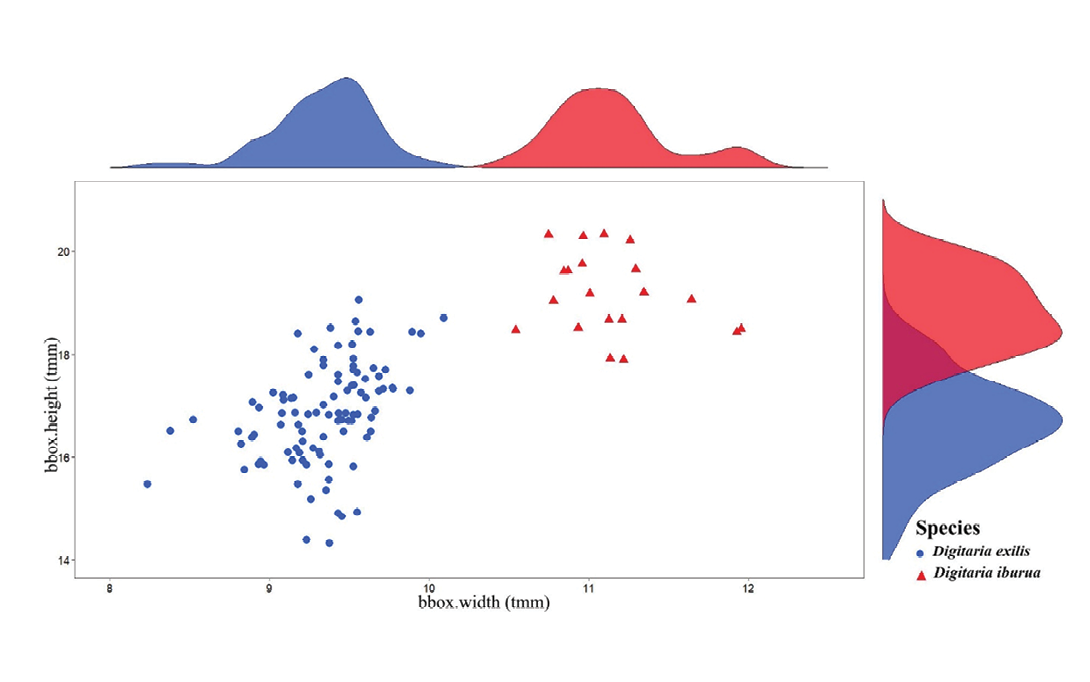
Discussion
This work sought to distinguish the two species of cultivated fonio based on the characteristics of their seeds in a context of genebank conservation. Seed morphometrics was used as a rapid, low-cost and non-destructive method to identify the two fonio species. Morphometrics has largely been used to describe and compare organism shapes, allowing reliable species identification. This was confirmed on organisms as varied as orchids (Chemisquy et al, 2009), mosquitoes (Chaiphongpachara et al, 2022), indigo plants (Soladoye et al, 2010), wheat species (Goriewa-Duba et al, 2018) and olive (Terral et al, 2004; Newton et al, 2014).
The tiny size of the seeds of both species is a source of practical difficulties, particularly for handling and visual identification. We showed that the Rigatoni R package (Rami, 2022) could be adapted to detect very small objects in images. While this package has been initially developed for the analysis of irregular shapes like peanut pods, we used here a smaller number of Rigatoni variables since fonio seeds have an oblong ellipsoid shape (Idu et al, 2008).
One consequence of small seed size is that seed lots can be heterogeneous due to the residual presence of undesirable biological material (pedicels, open glumes, foreign seeds), sand or stones in seed samples despite careful cleaning of the sample before measurement, as suggested by Koreissi-Dembélé et al (2013). A large number of seeds (over 200) per accession were scanned to ensure reliable results by limiting the influence of outliers and being able to detect them using robust quantitative methods. We implemented a fast method to remove extreme outliers from the analysis to make the identification more robust. Depending on the accession, between 0.4% and 8.8% of seeds were identified as extreme outliers.
White and black fonio are very similar as they share many agromorphological characteristics. According to Adoukonou-Sagbadja et al (2007), a clear-cut separation of both species was not possible using agromorphological traits such as plant height, number of tillers, leaf length, fresh and dry biomass weight, panicle length, and yield. This study focused on seed morphology and seed weight. We showed that seeds of D. iburua were significantly larger, heavier and had a more intense and darker brown colour than those of D. exilis. Seed width clearly distinguished D. exilis from D. iburua in our sample of accessions. Our results thus confirmed a difference in seed size between the two species, as previously noted by Echendu et al (2009) and Jideani (2012). The distinctiveness of D. exilis and D. iburua was also confirmed with respect to seed weight (Aliero et al, 2002; Nyam et al, 2017). We showed that seed area was a descriptor also separating the two species; however, we focused on seed width, a one-dimensional parameter whose variations are straightforward to interpret. It is worth noting that the differences between species, in seed size, weight and colour, were revealed despite the different conditions under which the accessions were collected and conserved. The species effect on these morphometric characteristics, therefore, appears stable, as it is greater than eventual environmental effects. Inversely, the more or less tapered shape of the seeds did not show a clear difference between species, but varied in an apparently structured way according to country of harvest (PCA, axis 2, Figure 6). The projection on a map (Supplemental Figure 5) of seed shape values (m.eccentricity descriptor cut into classes), at harvesting sites, visually confirmed that the most tapered D. exilis seeds came mainly from the northern edge of the sampling zone (Senegal, Mali, northern Burkina Faso, Niger), in drier climatic regions. This trend requires more in-depth studies in relation to climatic data.
The number of accessions used in our study differed markedly between the two species. This is partly because black fonio is predominantly grown in central Nigeria today, from where samples are available in genebanks. Moreover, some geographical origins could not be used for D. exilis because of incomplete passport data. We can't rule out the possibility that a more balanced sampling design, both in terms of species frequency and geographic distribution, might nuance our results, and that the species classification might be less categorical in a context of greater morphometric variability. However, our sampling design was based on genetic studies that already maximized the diversity and species geographical range in their sampling. Despite sampling constraints, PCA provided some confidence in the results. The first principal component (Figure 6), which differentiates the two species by seed size and colour saturation, did not appear to show any structure related to geographical origin. Further studies focusing on the effect of geographic origin on seed morphology should investigate this issue more precisely, both within and between species.
Our morphometric results also open up new avenues for research in archaeobotany. The morphometric approach proposed in this paper could be used to identify fonio in archaeological records, contributing to the reconstruction of the evolutionary history of both cultivated fonio and its wild relatives. Morphometrics of ancient seeds would provide a better understanding of the domestication and diffusion histories of D. exilis and D. iburua in West Africa. Indeed, morphometrics on crops allowed to confidently distinguish between domesticated versus wild forms and trace the evolution of cultivated forms through space and time, as in the case of grapevine (Bouby et al, 2013; Rôs et al, 2014; Bonhomme et al, 2021; Ucchesu et al, 2024) or date palm (Terral et al, 2012; Gros-Balthazard et al, 2016). Additional research with carbonization experiments is however needed to see the impact of charring on seed morphology in both species (Ivorra et al, 2024).
In order to preserve the existing fonio diversity from genetic erosion, germplasm collection and ex situ conservation is a necessity (Dansi et al, 2010). Correct taxonomic identification is all the more crucial for the plant genetic resources that are currently underrepresented in ex situ conservation facilities worldwide, as is the case for fonio, and therefore have a high priority for future collecting missions and urgent conservation measures (Guzzon et al, 2018). Labelling of accessions is essential to enhance the value of these collections and to make accessions usable. By enhancing passport data and combining it with additional information from other fields, new knowledge about plant genetic resources can be generated, which is crucial for the sustainable management of genebank collections.
In the case of fonio species, the seed width could be used as the sole criterion for a simple and inexpensive method to make their taxonomic identification and genebank passport data more reliable. This approach is particularly useful when genetic data are not available. As a new genebank conservation procedure, we suggest that the method proposed in this paper be applied to fonio accessions already conserved in genebank collections to ensure the reliability of fonio identification in passport data. It could also be systematically applied to any new fonio accession before its integration in genebank collections, especially for fonio originating from regions where both species are grown.
Supplemental Table 1. Sampling details: number of accessions by countries and species.
Supplemental Table 2. Information on the 118 accessions analyzed.
Supplemental Table 3. Results for the Principal Component Analysis carried out on the seven selected morphometric variables and the 118 fonio accessions.
Supplemental Figure 1. Histogram of median morphometric values by accession and TGW (thousand grain weight) for the two species.
Supplemental Figure 2. Position of the two fonio species, according to their median hue values, on the diagram representing a part of the sequence of Hue.
Supplemental Figure 3. Principal component analysis carried out on the seven selected morphometric variables and the 118 fonio accessions. Scree Plot: percentage of total variation explained by each principal component.
Supplemental Figure 4. Scatterplot with marginal distribution of seed area (x) and seed length (y) of the 118 fonio accessions.
Supplemental Figure 5. Projection on a map of seed shape values (m.eccentricity descriptor cut into classes).
Data and code availability statement
Dataset supporting the results of this article are available via Dataverse: https://dataverse.cirad.fr/dataset.xhtml?persistentId=doi:10.18167/DVN1/ZFZTWP
R script for the different analyses carried out throughout the paper are available on the CIRAD Gitlab platform: https://gitlab.cirad.fr/agap/fonio/seedfonio
Author contributions
SC, TK, AB, CB, and CL designed the research. EGAD, ARIBY, SS, YB, BMD, MCG, RYA, JAD, LA, MMB, EAU, HOO, SSI, SV, TK, AB and CB contributed to the sampling of the biological material or the curation of collections. SC generated the data. SC analyzed the data with inputs from CD, JFR, TK, AB, CB, and CL. SC and CL wrote the paper with substantial inputs from TK, AB, CD, CB.
Conflict of interest statement
The authors have no conflicts of interest to report.
Acknowledgments
The exploration of fonio diversity over the past decade has been facilitated by various research initiatives. These projects received funding from WAAPP/PPAAO 2A (CERA58ID06 SE), Agropolis Fondation (specifically the Agropolis Resource Center for Crop Adaptation and Diversity [ARCAD] project and the Cultivar project—ID 1504-007) under the Investissements d'Avenir programme (Labex Agro: ANR-10-LABX-0001-01) within the I-SITE MUSE framework (ANR-16-IDEX-0006). Additionally, support came from the French government ANR project (AfriCrop project, ANR-13-BSV7-0017) and the European Union Horizon 2020 research and innovation programme (EWA-BELT, 862848, ‘Linking East and West African farming systems experience into a BELT of sustainable intensification’).
We would also like to thank CRB GAMéT for providing us with seeds for our study (Centre de Ressource Biologique GAMéT, ARCAD – UMR AGAP Institut de Montpellier. https://doi.org/10.18167/infrastructure/00007)
References
Abrouk M., Ahmed H.I., Cubry P., Šimoníková D., Cauet S., Pailles Y., Bettgenhaeuser J., Gapa L., Scarcelli N., Couderc M., Zekraoui L., Kathiresan N., Čížková J., Hřibová E., Doležel J., Arribat S., Bergès H., Wieringa J.J., Gueye M., Kane N.A., Leclerc C., Causse S., Vancoppenolle S., Billot C., Wicker T., Vigouroux Y., Barnaud A., Krattinger S.G. (2020). Fonio millet genome unlocks African orphan crop diversity for agriculture in a changing climate. Nat Commun., 11, 4488. doi: https://doi.org/10.1038/s41467-020-18329-4
Adoukonou-Sagbadja, H., Wagner, C., Dansi, A., Ahlemeyer, J., Daïnou, O., Akpagana, K., Ordon F., Friedt W. (2007). Genetic diversity and population differentiation of traditional fonio millet (Digitaria spp.) landraces from different agro-ecological zones of West Africa. Theor. Appl. Genet., 115, 917-931. doi: https://doi.org/10.1007/s00122-007-0618-x
Adoukonou-Sagbadja, A.H. (2010). Genetic Characterization of Traditional Fonio Millets (Digitaria exilis, D. iburua STAPF) Landraces from West-Africa: Implications for Conservation and Breeding. PhD thesis, Justus-Liebig University, Giessen, Germany.
Aliero, A. A. & Morakinyo, J.A. (2002). Characterization of Digitaria exilis (Kipp) StapF and D. iburua Stapf Accessions. Nigerian Journal of Genetics, 16, 10-21. doi: https://doi.org/10.4314/njg.v16i1.42277
Animasaun, D. A., Awujoola, K. F., Oyedeji, S., Morakinyo, J. A., and Krishnamurthy, R. (2018). Diversity level of genomic microsatellite among cultivated genotypes of Digitaria species in Nigeria. African Crop Science Journal, 26(2), 305-313. doi: https://doi.org/10.4314/acsj.v26i2.11
Beck, J., Böller, M., Erhardt, A., Schwanghart, W. (2014). Spatial bias in the GBIF database and its effect on modeling species' geographic distributions. Ecological Informatics, 19, 10-15. doi: https://doi.org/10.1016/j.ecoinf.2013.11.002
Blench, R. (2016). Finger millet: the contribution of vernacular names towards its prehistory. Archaeol Anthropol Sci 8, 79–88. doi: https://doi.org/10.1007/s12520-012-0103-6
Bonhomme, V., Terral, J.F., Zech-Matterne, V., Ivorra, S., Lacombe, T., Deborde, G., Kuchler, P., Limier, B., Pastor, T., Rollet, P., Bouby, L. (2021) Seed morphology uncovers 1500 years of vine agrobiodiversity before the advent of the Champagne wine. Sci Rep., 11(1):2305. doi: https://doi.org/10.1038/s41598-021-81787-3
Bouby L., Figueiral I., Bouchette A., Rovira N., Ivorra S., Lacombe T., Pastor T., Picq S., Marinval P., Terral J.F. (2013) Bioarchaeological insights into the process of domestication of grapevine (Vitis vinifera L.) during Roman times in Southern France. PLoS One, 8(5): e63195. doi: https://doi.org/10.1371/journal.pone.0063195
Chaiphongpachara, T., Changbunjong, T., Sumruayphol, S., Laojun, S., Suwandittakul, N., Kuntawong K. (2022). Geometric morphometrics versus DNA barcoding for the identification of malaria vectors Anopheles dirus and An. baimaii in the Thai-Cambodia border. Sci Rep, 12, 13236. doi: https://doi.org/10.1038/s41598-022-17646-6
Chemisquy, M. A., Prevosti, F. J., and Morrone, O. (2009). Seed morphology in the tribe Chloraeeae (Orchidaceae): combining traditional and geometric morphometrics. Botanical journal of the Linnean Society, 160(2), 171-183. doi: https://doi.org/10.1111/j.1095-8339.2009.00968.x
Dansi, A., Adoukonou-Sagbadja, H., and Vodouhè, R. (2010). Diversity, conservation and related wild species of Fonio millet (Digitaria spp.) in the northwest of Benin. Genetic Resources and Crop Evolution, 57, 827-839. doi: https://doi.org/10.1007/s10722-009-9522-3
Echendu, C. A., Obizoba, I. C., Anyika, J. U., & Ojimelukwe, P. C. (2009). Changes in chemical composition of treated and untreated hungry rice “Acha” (Digitaria exilis). Pakistan journal of nutrition, 8(11), 1779-1785. doi: https://doi.org/10.3923/pjn.2009.1779.1785.
FAO. (2010). The Second Report on the State of the World’s Plant Genetic Resources for Food and Agriculture. Rome. url: https://www.fao.org/3/i1500f/I1500F.pdf
FAO. (2020). How the world's food security depends on biodiversity. Rome. url: https://www.fao.org/3/cb0416en/CB0416EN.pdf
Gepts, P. (2006). Plant Genetic Resources Conservation and Utilization: The Accomplishments and Future of a Societal Insurance Policy. Crop Science, 46, 2278-2292. doi : https://doi.org/10.2135/cropsci2006.03.0169gas
Goriewa-Duba, K., Duba, A., Wachowska, U., and Wiwart, M. (2018). An evaluation of the variation in the morphometric parameters of grain of six Triticum species with the use of digital image analysis. Agronomy, 8(12), 296. doi: https://doi.org/10.3390/agronomy8120296
Gros-Balthazard, M., Newton, C., Ivorra, S., Pierre, M. H., Pintaud, J. C., and Terral, J. F. (2016). The domestication syndrome in Phoenix dactylifera seeds: toward the identification of wild date palm populations. PloS ONE, 11(3), e0152394. doi: https://doi.org/10.1371/journal.pone.0152394
Grosjean, P., Ibanez, F., Etienne, M. (2018). pastecs: Package for Analysis of Space-Time Ecological Series. url: https://github.com/SciViews/pastecs
Guzzon, F., and Ardenghi, N.M.G. (2018). Could taxonomic misnaming threaten the ex situ conservation and the usage of plant genetic resources? Biodiversity and Conservation, 27, 1157-1172. doi: https://doi.org/10.1007/s10531-017-1485-7
Hema, D., and Kannan, D. S. (2019). Interactive color image segmentation using HSV color space. Sci. Technol. J, 7(1), 37-41. doi: https://doi.org/10.22232/stj.2019.07.01.05
Hunter D., Borelli T., Beltrame D.M.O., Oliveira C.N.S., Coradin L., Wasike V.W., Wasilwa L., Mwai J., Manjella A., Samarasinghe G.W.L., Madhujith T., Nadeeshani H.V.H., Tan A., Ay S.T., Güzelsoy N., Lauridsen N., Gee E., Tartanac F. (2019). The potential of neglected and underutilized species for improving diets and nutrition. Planta, 250, 709-729. doi: https://doi.org/10.1007/s00425-019-03169-4
Idu, M., J. U. Chokor, and O. Timothy. (2008). Effect of Various Hormones on the Germination of Fonio-Digitaria exilis L. International Journal of Botany, doi: https://doi.org/10.3923/ijb.2008.456.460
Ivorra, S., Tengberg, M., Bonhomme, V., Kaczmarek, T., Pastor, T., Terral, J.F., Gros-Balthazard, M. (2024). Leveraging the potential of charred archaeological seeds for reconstructing the history of date palm. Journal of Archaeological Science, 170, 106052. doi: https://doi.org/10.1016/j.jas.2024.106052.
Jideani, I.A. (2012). Digitaria exilis (acha/fonio), Digitaria iburua (iburu/fonio) and Eleusine coracana (tamba/finger millet) Non-conventional cereal grains with potentials. Scientific Research and Essays, 7, 3834-3843. doi: https://doi.org/10.5897/SRE12.416
Kaczmarek, T., Cubry, P., Champion, L., Causse, S., Couderc, M., Orjuela, J., ... & Leclerc, C. (2025). Independent domestication and cultivation histories of two West African indigenous fonio millet crops. Nature Communications, 16(1), 4067. doi: https://doi.org/10.1038/s41467-025-59454-2
Karl, K., MacCarthy, D., Porciello, J., Chimwaza,G., Fredenberg, E., Freduah, B.S., Guarin, J., Mendez Leal, E., Kozlowski, N., Narh, S., Sheikh, H., Valdivia, R., Wesley, G., Van Deynze, A., van Zonneveld, M., Yang, M.(2024). Opportunity Crop Profiles for the Vision for Adapted Crops and Soils (VACS) in Africa. doi: https://doi.org/10.7916/7msa-yy32
Koreissi-Dembélé, Y., Fanou-Fogny, N., Hulshof, P. J., and Brouwer, I. D. (2013). Fonio (Digitaria exilis) landraces in Mali: Nutrient and phytate content, genetic diversity and effect of processing. Journal of Food Composition and Analysis, 29(2), 134-143. doi: https://doi.org/10.1016/j.jfca.2012.07.010
Khoury, C.K., Bjorkman, A.D., Dempewolf H., Ramirez-Villegas J., Guarino L., Jarvis A., Rieseberg L.H., Struik P.C. (2014). Increasing homogeneity in global food supplies and the implications for food security. Proc Natl Acad Sci U S A, 111 (11), 4001-4006. https://doi.org/10.1073/pnas.1313490111
Lê, S., Josse, J. and Husson, F. (2008). FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software, 25(1). pp. 1-18. doi: https://doi.org/10.18637/jss.v025.i01
Mondini, L., Noorani, A., and Pagnotta, M. A. (2009). Assessing Plant Genetic Diversity by Molecular Tools. Diversity, 1(1), 19-35. doi: https://doi.org/10.3390/d1010019
Newton, C., Lorre, C., Sauvage, C., Ivorra, S., and Terral, J.-F. (2014). On the origins and spread of Olea europaea L. (olive) domestication: evidence for shape variation of olive stones at Ugarit, Late Bronze Age, Syria - a window on the Mediterranean Basin and on the westward diffusion of olive varieties. Vegetation History and Archaeobotany, 23(5), 567-575. doi: https://doi.org/s00334-013-0412-4
Nyam, D., Kwon-Ndung, E. and Ap, W. (2017). Genetic Affinity and Breeding Potential of Phenologic Traits of Acha (fonio) in Nigeria. Journal of Scientific and Engineering Research, 4(10), 91-101. url: https://irepos.unijos.edu.ng/jspui/handle/123456789/1966
Rami, J.F. (2022). Rigatoni: Object detection (typically grains) in images. R package version 0.9. url: https://github.com/jframi/rigatoni
Rôs, J., Evin, A., Bouby, L., and Ruas, M. P. (2014). Geometric morphometric analysis of grain shape and the identification of two-rowed barley (Hordeum vulgare subsp. distichum L.) in southern France. Journal of Archaeological Science, 41, 568-575. doi: https://doi.org/10.1016/j.jas.2013.09.015
Schloerke B., Cook D., Larmarange J., Briatte F., Marbach M., Thoen E., Elberg A., Crowley J. (2024). GGally: Extension to 'ggplot2'. R package version 2.2.1. url: https://ggobi.github.io/ggally/
Soladoye, M. O., Sonibare, M. A., and Chukwuma, E. C. (2010). Morphometric Study of the Genus Indigofera Linn. (Leguminosae-Papilionoideae) in South-Western Nigeria. International Journal of Botany, 6: 343-350. doi: https://doi.org/10.3923/ijb.2010.343.350
Stamp, P., Messmer, R. and Walter, A. (2012), Competitive underutilized crops will depend on the state funding of breeding programmes: an opinion on the example of Europe. Plant Breeding, 131: 461-464. doi: https://doi.org/10.1111/j.1439-0523.2012.01990.x
Terral, J. F., Alonso, N., Capdevila, R. B. I., Chatti, N., Fabre, L., Fiorentino, G., Marinval, P., Pérez-Jordà, G., Pradat, B., Rovira, N., Paul, A. (2004). Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. Journal of Biogeography, 31. 63-77. doi: https://doi.org/10.1046/j.0305-0270.2003.01019.x
Terral, J.F., Newton, C., Ivorra, S., Gros-Balthazard, M., Morais, C., Picq, S., Tengberg, M., Pintaud, J.C. (2012). Insights into the historical biogeography of the date palm (Phoenix dactylifera L.) using geometric morphometry of modern and ancient seeds. Journal of Biogeography, 39. 929-941. doi: https://doi.org/10.1111/j.1365-2699.2011.02649.x
Ucchesu, M., Depalmas, A., Sarigu, M., Gardiman, M., Lallai, A., Meggio, F., Usai, A., Bacchetta, G. (2024). Unearthing Grape Heritage: Morphological Relationships between Late Bronze–Iron Age Grape Pips and Modern Cultivars. Plants, 13(13): 1836. doi: https://doi.org/10.3390/plants13131836
Ulian, T., Diazgranados M., Pironon S. et al (2020). Unlocking plant resources to support food security and promote sustainable agriculture. Plants, People, Planet, 2: 421-445. doi: https://doi.org/10.1002/ppp3.10145
Weise, S., Lohwasser, U., Oppermann, M. (2020). Document or Lose It - On the Importance of Information Management for Genetic Resources Conservation in Genebanks. Plants 2020, 9, 1050. doi: https://doi.org/10.3390/plants9081050
Zeileis, A., Fisher, J. C., Hornik, K., Ihaka, R., McWhite, C. D., Murrell, P., Stauffer, R., and Wilke, C. O. (2020). colorspace: A Toolbox for Manipulating and Assessing Colors and Palettes. Journal of Statistical Software, 96(1), 1-49. doi: https://doi.org/10.18637/jss.v096.i01